Pathogenic bacteria are highly specialized micro-organisms that are a leading cause of human, livestock and crop disease, resulting in significant health and economic loss worldwide. How these pathogens, which sometimes even live inside the cells of their hosts, once evolved is poorly understood.
An international team of researchers has now found how bacterial pathogens evolved. Their work, published in Nature Microbiology today, reveals that the transition from free-living ancestors to modern day pathogens progressed in a stepwise manner—involving gene gain and loss events, but also the repurposing of existing genes.
While most micro-organisms are benign or even essential to sustain life, some bacteria are known to cause disease in organisms like humans, animals and plants. Intracellular pathogens can establish persistent and sometimes lifelong infections. While bacterial pathogens are known to have evolved multiple times independently, not much is known about their evolutionary origin. The new research published in Nature Microbiology reveals how Rickettsiales, a group of obligate intracellular bacteria that includes several notorious pathogens of humans and cattle, evolved from once free-living bacteria.
Aquatic relatives of bacterial pathogens
While studying the diversity of micro-organisms in oceanic waters, an international group of researchers discovered several new species of bacteria that were distantly related to Rickettsiales. This group includes a wide variety of obligate intracellular bacteria—one of which is the causative agent of human epidemic typhus, Rickettsia prowazekii. Other Rickettsiales, such as Wolbachia, are known to infect most insects worldwide. “To find distant relatives of Rickettsiales in oceanic waters was somewhat surprising,” says Max Emil Schön from Uppsala University in Sweden and co-lead author of the study. “We wondered if these bacteria, like Rickettsiales, were obligate intracellular pathogens, or whether they were part of the free-living bacterioplankton.”
The researchers discovered these new bacteria by mining metagenomic data—obtained by analyzing genetic material of all organisms that live in an environment—which doesn’t rely on growing organisms in the lab. “The majority of microbial life on Earth currently can’t be grown in the lab,” explains Thijs Ettema, professor in Microbiology at Wageningen University & Research in The Netherlands who led the work. “Using metagenomic data, we managed to reconstruct the genomes of these new oceanic bacteria. By analyzing their genomes, we could predict that their lifestyles were very different from the previously known obligate intracellular Rickettsiales.”
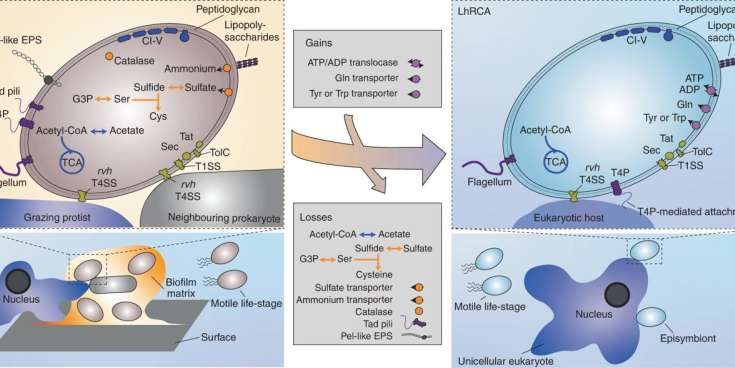
Thijs Ettema’s team noticed that the genomes of these new Rickettsiales encoded many genes that were absent from their host-associated and pathogenic relatives. “Besides several metabolic genes typically absent from Rickettsiales, we found genes involved in motility, surface attachment and the formation of biofilms. The presence of such genes points at a free-living lifestyle,” says co-lead author Joran Martijn, former member of Ettema’s lab who currently works at Dalhousie University in Canada.
Genes typically involved in pathogenic lifestyles, such as those involved in energy parasitism or host cell manipulation, were not identified. The researchers did however find genes encoding a so-called “type 4 secretion system,” a microscopic needle-like structure used by bacteria to interact with host cells. “We speculate that this system is not necessarily used to interact with host cells like other Rickettsiales, but rather to kill competing bacteria, or to fence off predatory microbes,” Martijn adds.
Stepwise evolution of pathogenic lifestyle
By comparing the genomes of the newly discovered species with those of previously known Rickettsiales, the team of researchers managed to reconstruct the evolution of host association and pathogenicity in the Rickettsiales. “We suggest that the free-living ancestor of Rickettsiales repurposed its needle-like type 4 secretion system to interact with and manipulate host cells,” Ettema speculates.
“Subsequently, many metabolic genes and genes affiliated with free-living lifestyle were lost as the ancestral Rickettsiales became more dependent on its host for metabolites and energy. This was then mirrored by the acquisition of genes involved in host manipulation and energy parasitism,” he adds. The team hopes to be able to grow the newly identified species in the lab in the future, aiming to further unravel the evolutionary pathway towards host association and pathogenicity in Rickettsiales.
#PathogenicBacteria